Accueil du site > Production scientifique > Structure of sodiated octaglycine : IRMPD spectroscopy and molecular modeling
Structure of sodiated octaglycine : IRMPD spectroscopy and molecular modeling
Date de publication: 1er février 2010
D. Semrouni, O. P. Balaj, F. Calvo, C. Correia, C. Clavaguera, G. Ohanessian
J. Am. Soc. Mass Spectrom. 21, 728 (2010). DOI
Travail réalisé sur le site de l’Université de Paris-Sud.
Abstract
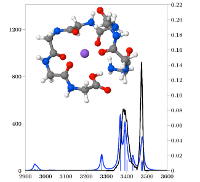
The structure of the sodiated peptide GGGGGGGG-Na^+ or G_8 -Na^+ was investigated by InfraRed Multiple Photon Dissociation (IRMPD) spectroscopy and a combination of theoretical methods. IRMPD was carried out in both the fingerprint and N-H/O-H stretching regions. Modeling used the polarizable force field AMOEBA in conjunction with the Replica-Exchange Molecular Dynamics (REMD) method, allowing an efficient exploration of the potential energy surface. Geometries and energetics were further refined at B3LYP-D and MP2 quantum chemical levels. The IRMPD spectra indicate that there is no free C-terminus OH and that several N-H’s are free of hydrogen bonding while several others are bound, however not very strongly. The structure must then be either of the charge solvation (CS) type with a hydrogen-bound acidic OH, or a salt bridge (SB). Extensive REMD searches generated several low energy structures of both types. The most stable structures of each type are computed to be very close in energy. The computed energy barrier separating these structures is small enough that G_8 -Na^+ is likely fluxional with easy proton transfer between the two peptide termini. There is however good agreement between experiment and computations in the entire spectral range for the CS isomer only, which thus appears to be the most likely structure of G_8 -Na^+ at room temperature.








