Accueil du site > Production scientifique > Structural and Energetic Effects of O2′-Ribose Methylationof Protonated Pyrimidine Nucleosides
Structural and Energetic Effects of O2′-Ribose Methylationof Protonated Pyrimidine Nucleosides
Date de publication: 21 août 2019
C. C. He ; L. A. Hamlow ; Y. Zhu ; Y. W. Nei ; L. Fan ; C. P. McNary ; P. Maitre ; V. Steinmetz ; B. Schindler ; I. Compagnon ; P. B. Armentrout ; M. T. Rodgers
J. Am. Soc. Mass Spectrom. 30 (11) 2318-2334 (2019). DOI
Travail réalisé sur le site de l’Université Paris Sud.
Abstract
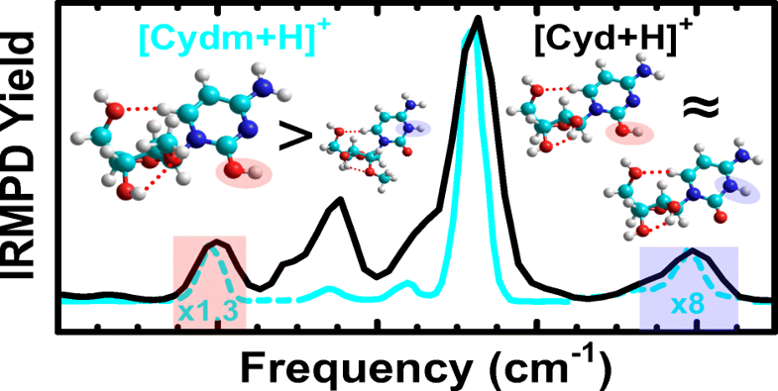
The 2 ’-substituents distinguish DNA from RNA nucleosides. 2 ’-O-methylation occurs naturally in RNA and plays important roles in biological processes. Such 2 ’-modifications may alter the hydrogen-bonding interactions of the nucleoside and thus may affect the conformations of the nucleoside in an RNA chain. Structures of the protonated 2 ’-O-methylated pyrimidine nucleosides were examined by infrared multiple photon dissociation (IRMPD) action spectroscopy, assisted by electronic structure calculations. The glycosidic bond stabilities of the protonated 2 ’-O-methylated pyrimidine nucleosides, [Nuom+H](+), were also examined and compared to their DNA and RNA nucleoside analogues via energy-resolved collision-induced dissociation (ER-CID). The preferred sites of protonation of the 2 ’-O-methylated pyrimidine nucleosides parallel their canonical DNA and RNA nucleoside analogues, [dNuo+H](+) and [Nuo+H](+), yet their nucleobase orientation and sugar puckering differ. The glycosidic bond stabilities of the protonated pyrimidine nucleosides follow the order : [dNuo+H](+) < [Nuo+H](+) < [Nuom+H](+). The slightly altered structures help explain the stabilization induced by 2 ’-O-methylation of the pyrimidine nucleosides.








